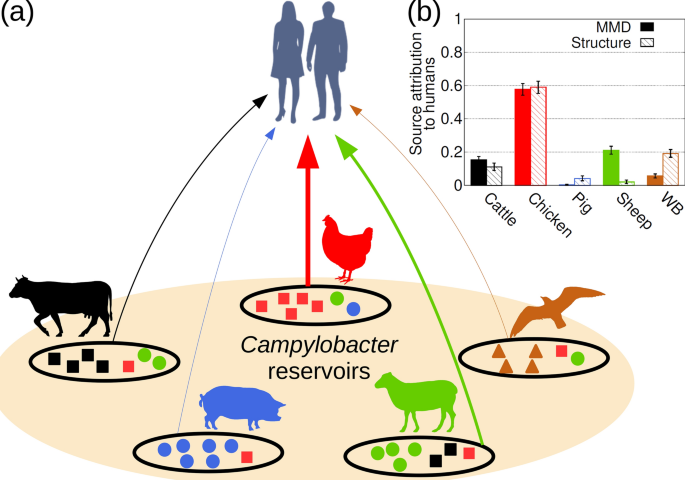
Scientists at the University of Aberdeen in Scotland (Departments of Physics and Biological Sciences) have developed a method that could help to identify the source of food poisoning. They developed a minimal multilocus distance (MMD) method which rapidly deals with large data sets such as whole genome sequence (WGS) well as methods for optimally selecting loci. The MMD method can be used to train the computer to identify likely sources of origin of a Campylobacter infection with high speed and accuracy. The methods are generic, easy to implement for WGS and proteomic data and have wide application. It is wise to employ a number of methods on each dataset to decide which set of loci are optimal. The performance of different locus selection strategies can be tested relatively fast with the MMD method. @ https://www.nature.com/articles/s41598-020-68740-6?proof=t
Machine learning, mining whole genome sequencing to find sources of food poisoning
Whole genome sequence (WGS) data could transform our ability to attribute individuals to source populations. However, methods that efficiently mine these data are yet to be developed. We present a minimal multilocus distance (MMD) method which rapidly deals with these large data sets as well as methods for optimally selecting loci. This was applied on WGS data to determine the source of human campylobacteriosis, the geographical origin of diverse biological species including humans and proteomic data to classify breast cancer tumours. The MMD method provides a highly accurate attribution which is computationally efficient for extended genotypes. These methods are generic, easy to implement for WGS and proteomic data and have wide application.