Researchers from North Carolina State University (Department of Population Health and Pathobiology, College of Veterinary Medicine) published an article in Front. Vet. Sci., 01 September 2021. The researchers describe the genome sequencing of a virulent Salmonella Enteritidis (SE) strain that sickened two poultry flocks in consecutive years and found that it was antibiotic-resistant and could potentially infect humans. In a broiler embryo lethality assay, the SE strain designated SE_TAU19 was characterized through genome sequencing for antimicrobial susceptibility and virulence. SE_TAU19 was resistant to nalidixic acid and sulfadimethoxine. The strain was virulent to embryos with 100% mortality of all challenged broiler embryos within 3.5 days. Screening the SE_TAU19 whole-genome sequence revealed seven antimicrobial resistance (AMR) genes, 120 virulence genes, and two IncF plasmid replicons corresponding to a single, serovar-specific pSEV virulence plasmid. This study revealed that the quinolone and sulfonamide resistance of the SE_TAU19 strain was lethal to broiler embryos and harbored a pSEV plasmid and typical SE virulence genes. The SE_TAU19 strain is deadly to poultry, antibiotic-resistant, and infectious, but it could also infect humans. @ https://www.frontiersin.org/articles/10.3389/fvets.2021.725737/full
Genomic Characterization of Salmonella Enteritidis Strain Causing Persistent Infections in Broiler Chickens
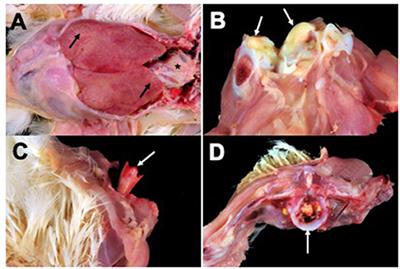
Virulent strains of Salmonella enterica subsp. enterica serovar Enteritidis (SE) harbored by poultry can cause disease in poultry flocks and potentially result in human foodborne illness. Two broiler flocks grown a year apart on the same premises experienced mortality throughout the growing period due to septicemic disease caused by SE. Gross lesions predominantly consisted of polyserositis followed by yolk sacculitis, arthritis, osteomyelitis, and spondylitis. Tissues with lesions were cultured yielding 59 SE isolates. These were genotyped by Rep-PCR followed by whole-genome sequencing of 15 isolates which were clonal. The strain, SE_TAU19, was further characterized for antimicrobial susceptibility and virulence in a broiler embryo lethality assay. SE_TAU19 was resistant to nalidixic acid and sulfadimethoxine and was virulent to embryos with 100% mortality of all challenged broiler embryos within 3.5 d. Screening the SE_TAU19 whole-genome sequence revealed 7 antimicrobial resistance genes, 120 virulence genes, and 2 IncF plasmid replicons corresponding to a single, serovar-specific pSEV virulence plasmid. The pef, spv, and rck virulence genes localized to the plasmid sequence assembly. We report phenotypic and genomic features of a virulent SE strain from persistently infected broiler flocks and present a workflow for SE characterization from isolate collection to genome assembly and sequence analysis. Further SE surveillance and investigation of SE virulence in broiler chickens is warranted.