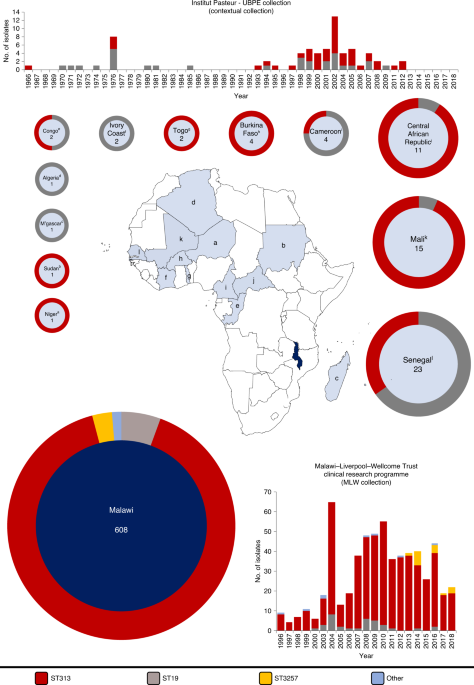
A new article (Pulford, C.V., Perez-Sepulveda, B.M., Canals, R. et al. entitled “Stepwise evolution of Salmonella Typhimurium ST313 causing bloodstream infection in Africa”, Nat Microbiol (2020), published 21 December 2020), explores how Salmonella evolved to kill hundreds of thousands of immunocompromised people in Africa. Bloodstream infections caused by a drug-resistant type of Salmonella Typhimurium ST313 are a significant public health concern in Africa, causing ~49,600 deaths every year. In the study, mutations that influenced gene function during the evolution of ST313 were identified for the first time. The researchers also discovered a new antibiotic-susceptible lineage of ST313 that emerged in Malawi in 2016 and is closely related to Salmonella variants that cause stomach infections in the United Kingdom and Brazil. The study provides a snapshot of the extensive repertoire of genomic changes that shaped the emergence of successful ST313 lineages. The researchers speculate that changes in antibiotic (the phased removal of chloramphenicol from clinical practice) in Malawi between 2002 and 2015 could have resulted in the emergence of this new antibiotic-susceptible ST313 lineage. @ https://www.nature.com/articles/s41564-020-00836-1#Sec2
Evolution of Salmonella Typhimurium ST313 and how it made the leap from gut to the bloodstream
Stepwise evolution of Salmonella Typhimurium ST313 causing bloodstream infection in Africa
Stepwise evolution of invasive Salmonella Typhimurium in Africa is defined using genotypic and phenotypic analyses of isolates collected over a 50-yr period.
No comments