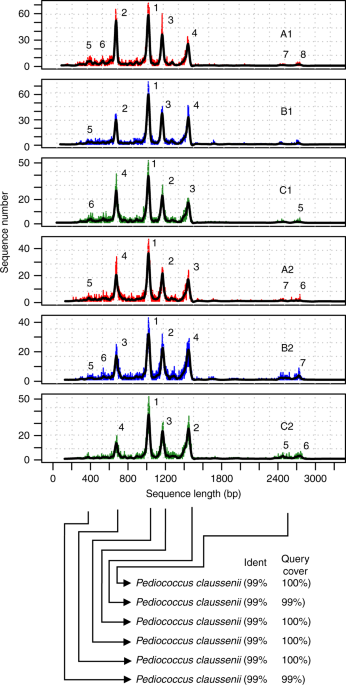
The process of fast and accurate bacterial identification, sub-typing, and strain-level differentiation is of high importance in epidemiology. A research group from Copenhagen, Denmark, published an article describing a rapid, accurate and inexpensive method for the identification of bacterial isolates down to species/strain level. The new method developed for bacterial DNA enrichment and tagmentation allowing fast (<24 h) and cost-effective species-level identification and strain level differentiation using the MinION portable sequencing platform (ON-rep-seq). The method in combination with ONT-sequencing platforms, allows for highly cost-effective, bulk screening of bacterial isolates with species and strain-level resolution. The method is claimed to have the potential to become a modern standard molecular-based method with multiple applications in research, industry, and medicine. @ https://www.nature.com/articles/s42003-019-0617-x#Sec7
A rapid method for species-level identification and strain-level differentiation
DNA enrichment and tagmentation method for species-level identification and strain-level differentiation using ON-rep-seq
Åukasz Krych et al. uses MinION portable sequencing platform to develop a fast and cost-effective method for bacterial DNA enrichment and tagmentation (ON-rep-seq). Their method allows species-level identification and strain-level differentiation of 288 isolates on a single flow for less than $2 per sample at very high speed.
No comments