Cornell University scientists create a national atlas for Listeria monocytogenes collected from soils across the US
 Dr. Martin Wiedmann, Jingqiu Liao, and their team from Cornell University created an atlas of Listeria monocytogenes in US soils. The research was published in Nature Microbiology (Jingqiu Liao, Xiaodong Guo, Daniel L. Weller, Shaul Pollak, Daniel H. Buckley, Martin Wiedmann, Otto X. Cordero. Nationwide genomic atlas of soil-dwelling Listeria reveals effects of selection and population ecology on pangenome evolution. Nature Microbiology, 2021; 6 (8): 1021).
Dr. Martin Wiedmann, Jingqiu Liao, and their team from Cornell University created an atlas of Listeria monocytogenes in US soils. The research was published in Nature Microbiology (Jingqiu Liao, Xiaodong Guo, Daniel L. Weller, Shaul Pollak, Daniel H. Buckley, Martin Wiedmann, Otto X. Cordero. Nationwide genomic atlas of soil-dwelling Listeria reveals effects of selection and population ecology on pangenome evolution. Nature Microbiology, 2021; 6 (8): 1021).
The study developed a nationwide atlas of 1,854 Listeria isolates, collected systematically from soils across the United States.
Initially, the Cornell team requested hundreds of other scientists across the country to collect soil samples from undisturbed places in the natural world, such as the off-trail areas of state and national parks. Team members collected additional soil samples. From the samples obtained, they developed a nationwide atlas of 1,854 Listeria isolates.
The Cornell team found that Listeria was present in a variety of environments. Listeria presence was affected by soil moisture, molybdenum, and salinity concentrations.
When a researcher identifies a Listeria, the atlas will inform them where the Listeria resides in the USA. Identifying the location can pinpoint the source of the organism found in ingredients processing facilities and finished products.
Whole-genome data from 594 Listeria representative strains allowed the team to break down the Listeria strains into 12 families, or phylogroups, each with differences in habitat breadth and being native to a single defined geographical location (endemism). “Cosmopolitan” phylogroups, prevalent across many different habitats, had more open pangenomes and displayed weaker linkage disequilibrium, reflecting higher rates of gene gain and loss and allele exchange than phylogroups with narrow habitat ranges.
Distribution of each Listeria phylogroup across the US (https://www.nature.com/articles/s41564-021-00935-7)
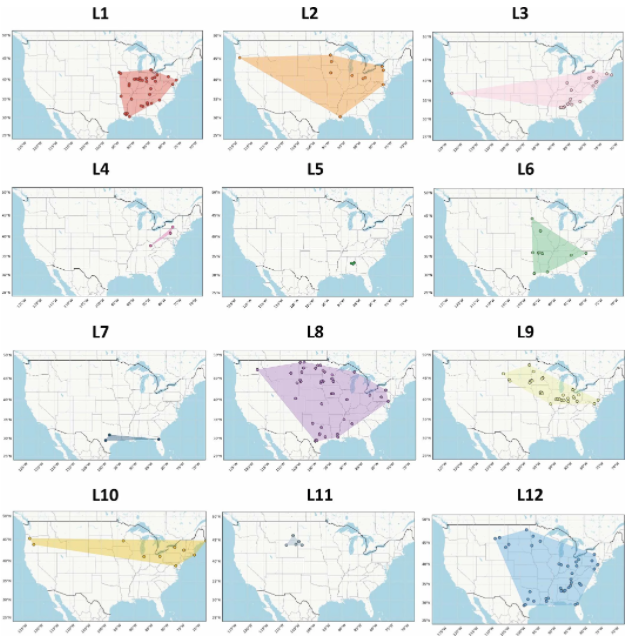
Cosmopolitan phylogroups also had a significant fraction of genes affected by positive selection. The effect of positive selection was more pronounced in the phylogroup-specific core genome, suggesting that lineage-specific core genes are essential drivers of adaptation. These results indicate that genome flexibility and recombination are the consequence of selection to survive in variable environments.
The Cornell news reported that senior author Dr. Martin Wiedmann said that the group is trying to determine the risk of getting Listeria from soil and different locations. The team created a more systematic way of assessing how frequently different Listeria are found in different locations. He also said that it was difficult to compare and assess Listeria diversity in different locations before this atlas was created.
Dr. Jingqiu Liao, the other senior author, said, “With whole genome sequencing and comprehensive population genomics analyses, we provided answers to the ecological and evolutionary drivers of bacterial genome flexibility – an important open question in the field of microbiology.” She said that the work can serve as a reference for future population genomics studies and will likely benefit the food industry by locating Listeria contaminations that may have a natural origin.
The atlas will tell scientists where Listeria and other related species reside within the US, which could help them trace and pinpoint sources of Listeria found in ingredients, food processing facilities, and finished products.
