CDC Report on Incidence and Trends of Infections with Pathogens Transmitted Through Food for 2016
 The CDC report describes surveillance data for 2016 nine pathogens (Campylobacter, Cryptosporidium, Cyclospora, Listeria, Salmonella, Shiga toxin-producing Escherichia coli (STEC), Shigella, Vibrio, and Yersinia). The data of 2016 is compared with the data from 2013-2015.
The CDC report describes surveillance data for 2016 nine pathogens (Campylobacter, Cryptosporidium, Cyclospora, Listeria, Salmonella, Shiga toxin-producing Escherichia coli (STEC), Shigella, Vibrio, and Yersinia). The data of 2016 is compared with the data from 2013-2015.
FoodNet identified 24,029 infections, 5,512 hospitalizations, and 98 deaths caused by these pathogens. The data include both incidents confirmed by culturing the organisms and culture-independent diagnostic tests (CIDTs). CIDT can identify (by rapid diagnostic tests) the general type of bacteria causing illness within hours, without having to culture or grow the bacteria in a laboratory.
This is the first time that the CDC report also includes CIDTs (total number of infections from bacterial infections diagnosed only by rapid diagnostic tests). Previously, the report counted foodborne bacterial infections confirmed only by traditional culture-based methods of total numbers.
Without a bacterial isolate from the culturing process, public health scientists can’t perform tests that determine an organism’s strain or subtype (such as DNA fingerprints), resistance pattern, or other characteristics.
In this report confirmed bacterial infections are defined as isolation of the bacterium from a clinical specimen by culture. In a “CIDT positive–only” bacterial infection was positive only by CIDT result that was not confirmed by culturing.
Results and Comparison
Annual data from the CDC’s Foodborne Diseases Active Surveillance Network (FoodNet) show that Campylobacter and Salmonella caused the most reported bacterial foodborne illnesses in 2016.
The largest number of confirmed or CIDT positive–infections in 2016 was reported for Campylobacter (8,547), followed closely by Salmonella (8,172). Shigella had 2,913 cases, and STEC 1,845. However, the largest number of confirmed positive infections was reported for Salmonella, followed by Campylobacter.
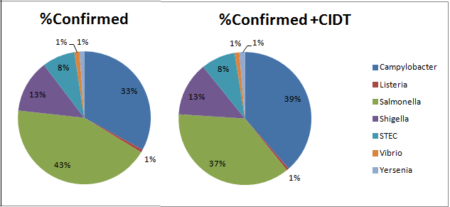
The proportion of infections that were CIDT positive without culture confirmation in 2016 was largest for Campylobacter (32%) and Yersinia (32%). There was a significant increase in CIDT in 2016 over 2013-2015.
Compared with 2013–2015, the 2016 incidence of Campylobacter infection was significantly lower (11% decrease) when only confirmed infections were included, yet it was not significantly different when including confirmed or CIDT positive–only infections. Incidence of STEC infection was significantly higher for confirmed infections (21% increase) and confirmed or CIDT positive–only infections (43% increase).
Among 7,554 confirmed Salmonella cases in 2016, serotype information was available for 6,583 (87%) of the cases. The most common serotypes were Enteritidis (17%), Newport (11%), and Typhimurium (9%). Salmonella Typhimurium infections decreased 18% in 2016 compared with the average for 2013-2015. This is perhaps due to the immunization of chicken flocks by producers.
Discussion Points
When including both culture confirmed with non culture confirmed, the incidence rates in 2016 were higher for each of the pathogens over previous years. This was mainly due to the increase in non- cultured CIDTs.
For example, the incidence of confirmed Campylobacter infections in 2016 was significantly lower than the 2013–2015 average. However, when including the non-cultured CIDT positive infections, a slight but not significant increase occurred. Similarly, the increase in STEC incidence is driven by the increase in STEC non-O157, which is not typically included in routine stool culture testing since it requires specialized methods. As a result, interpreting the incidence data is more complex due to the changing diagnostic landscape with unknown changes in frequency of testing, varying test performance, and decreasing availability of isolates for sub typing.
The new data reflect the increasing popularity of rapid tests known as culture-independent diagnostic tests, or CIDTs. According to the report, positive results from the rapid tests can be followed by culturing the organisms. However, often it is not done. The CIDT faster results have immediate benefit for treatment. Nevertheless, it does not collect needed data on strains and their antibiotic resistance.
The change from culture methodology to CIDT creates new problem comparing data from this year to previous years. Changes in the number of new infections could reflect changes in testing practices rather than a true increase in infections. Therefore, comparisons of the 2016 data with data from previous years may not accurately reflect trends. Estimated infections in 2016 and in years past are accurate, but cannot be directly compared.
