A Novel Concentration Device for the Detection of Food Pathogens
 Nature published in July 2017, an article byGwangseong Kim, Horatiu Vinerean & Angelo Gaitas on a simple novel device to concentrate and detect food pathogens (immunocapturing method). The technique has the potential of being used for both clinical applications and food testing.
Nature published in July 2017, an article byGwangseong Kim, Horatiu Vinerean & Angelo Gaitas on a simple novel device to concentrate and detect food pathogens (immunocapturing method). The technique has the potential of being used for both clinical applications and food testing.
System Set-Up
The technique employs polymer (polydimethylsiloxane) tubes (1.02 mm in diameter) coated with an antibody. The test sample is circulating through the antibody coated tubes. The re-circulating of liquid media containing the bacteria through the antibody conjug ated tubes result in the capturing of the pathogens by the conjugated antibodies.
ated tubes result in the capturing of the pathogens by the conjugated antibodies.
Several tubes can be used with different antibodies in each, thereby allowing the capture of different pathogens. Alternatively, several identical tubes can be used to increase the efficiency of the capturing.
As a result, the pathogens present in the sample are concentrated and accumulated in the tubes. This concentration step results in a higher concentration of the pathogens in a small volume of liquid.
Results
The results show that in larger volumes of 100-250 mL and small starting bacterial numbers of anywhere from 1 to 10 CFU anywhere from 55%-91% of bacteria were captured inside the tubes within 6-7 hours.
Ground chicken and ground beef were used as matrices to demonstrate the ability of the immuno-capturing method. 25 CFU of Salmonella typhimurium in 25 grams of ground meat was used to show the systems ability to work with real foods. The product was diluted 1:10 in 225 ml of buffered peptone water (BPW) or Romer Labs Primary enrichment media supplemented with phage. After 5-7 hours Salmonella was detected from these samples, representing significant time savings over the traditional methodology.
The two food matrices tested did not clog the 1mm tubes. To test larger volumes of samples required in food pathogens, long (120 cm) antibody coated tube was split into four 30 mm tubes. The 250 ml sample was circulated approximately 10 times in the 7-hour experiment.
Use of Molecular Methods: The S. Typhimurium DNA was directly extracted from the concentration tubes by inserting DI water in the tube and heating to 100 °C for 10 minutes. Other methods for DNA extraction were also tested. Detection of the presence of the pathogens was done using either microscope fluorescence imaging or RT PCR. 10 µm from the content can be directly used for RT PCR without further purification steps.
Use of Lateral flow devices: have a higher limit of detection than PCR, and therefore requires longer enrichment time. However, they are low cost and easy to use. Therefore they also were tested with the immunocapturing method.
As shown below, 25 cfu of S. typhimurium in 25 gram of ground meat could detect in 14 hours with traditional enrichment, and in 9 hours when using the Romer Primary enrichment medium with phage. These time frames are significantly lower than the traditional methodology (36-44 hours).
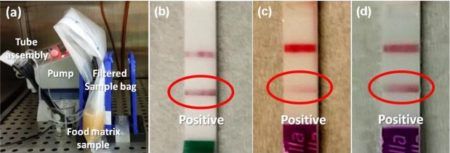
(b) Positive results using Neogen Reveal 2.0 Salmonella strip in 14 hours in non-selective media.(c) Positive result using Romer Labs RapidChek SELECT Salmonella strip in 14 hours in non-selective media, (d) Positive result using Romer Labs RapidChek SELECT Salmonella strip in 9 hours in selective media
Bottom-line
There is certainly a need for a faster method to find food pathogens because it allows for faster intervention and faster corrective action. It allows to link pathogen strains to specific cases and can be useful in preventing outbreaks and illnesses.
This novel method can allow for results from food matrices in less than a single shift. However, the technology is currently in prototype stage and will need to be developed to a full commercial product.
The inventors of the technology are currently seeking funding to finish the commercialization of the product. They expect the product to be commercially available in the next two years.
